Case study: Preference for the TIM-Barrel Fold among GroEL Substrates
View as Movie  BeanShell
script along with data (zipped)
BeanShell
script along with data (zipped) 
Keywords:
nominal (or categorical) comparisons, external data, generic XML format
Initial situation:
Substrates depending on the GroEL chaperonin were identified in E.coli.
Question:
- Do GroEL substrates prefer certain structural folds?
Data:
We used generic XML files that contain SCOP fold assignments that were taken from the public PEDANT database groel_substrates_ecoli at jerboas.gsf.de that can be accessed by http://pedant.gsf.de. As assignment threshold we used 1E-4. All folds assignments that were better than this cutoff were taken under consideration. For a more sensitive and exhaustive SCOP fold homology modelling and analysis, please refer to the publication of Kernel et al. (2005) Cell.
| File | Content |
| groel_substrates_ecoli.xml | All substrates of GroEL (class 1,2 & 3) in E.coli with SCOP fold assignments |
| Escherichia_coli_K12_updated.xml | Fold assignments of the whole E.coli K12 proteome |
Steps
Step 1: Data import
Simply import the 2 protein sets to PROMPT by using the Generic XML import feature. Choose “Import -> Generic XML Annotations -> Generic XML File”
Step 2: Analysis & Results
Select both input entries and choose from the menu:
"Analyze -> Generic Annotations -> Compare annotations between 2 sets -> Symbolic feature comparison"
Then select in both sets the same symbolic feature that should be compared (here we select ScopFold). As result all categories (here folds) that were found only in one set were returned. Additionally, the fold frequency differences were returned; this result entry is labelled Compare:symbolic:enrichment. Select this entry and use the right mouse click to open the context menu and afterwards use Visualise to show a plot.
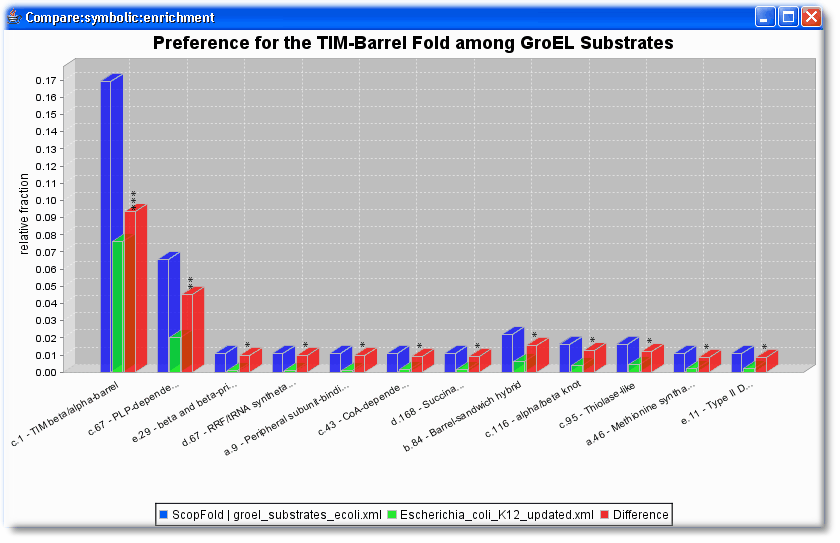
Figure 1: Frequency of SCOP folds in GroEL substrates compared with the whole E.coli proteome. Only folds that were found at least two times in both sets and were significantly different at the significance level of 0.05.
Interestingly, substantially enriched among the GroEL substrates are proteins with TIM-barrel domains. We suggest that the chaperonin system may have facilitated the evolution of this fold into a versatile platform for the implementation of numerous enzymatic functions. Kerner et. al (2005) Cell.
As seen in this example, PROMPT can compare the frequency of any nominal annotations (like SCOP folds here) and calculates whether the observed frequency differences are statistically significant. The p-values (*<0.05, **<0.01, *** <0.001) are indicated on top of the red bars.
Tip: You can easily change the order of the categories plotted. For example to sort by the p-value, open the data in the spread sheet viewer and save the sorted results as a text file. Then load the sorted result file in PROMPT again by using the "Load data" option of the right context menu in PROMPT's result area.
Summary:
- PROMPT can compare any nominal or categorical data. In the example here we used SCOP folds.
- The Generic XML file or the tab delimited input format allows the import of any kind of nominal or numeric data.
- PROMPT tests for statistical significance automatically
- The ready-to-use visualisations can be easily adapted to one's own needs. Here in the example we sorted by significance.
More:
Start PROMPT, Download PROMPT or sign up to the Community Mailing List
Previous case study: |
Back to the Case studies Overview |
Next case study: Thermophilic vs. mesophilic organism |
