Case study: Map a list of GenBank identifiers with a list of Swiss-Prot codes
View as Movie
 BeanShell
script along with data (zipped)
BeanShell
script along with data (zipped) 
Keywords:
id mapping, automatic sequence retrieval, GenBank, Swiss-Prot [IDs vs. IDs using BLAST or FASTA34]
Initial situation:
You have a list of GenBank identifiers and a list of Swiss-Prot accessions. Both list represent the same protein set. Now you want to find the corresponding identifiers.
Data:
We use a text file with GenBank IDs and one with Swiss-Prot codes.
| File | Content |
| genbank_ids.txt | Text file with GenBank identifiers |
| sprot_ids.txt | Text file with Swiss-Prot codes |
Steps
Step 1: Data import
The file with the GenBank identifier can be imported by choosing “Import -> GenBank -> GenBankIds” from PROMPT's menu line.
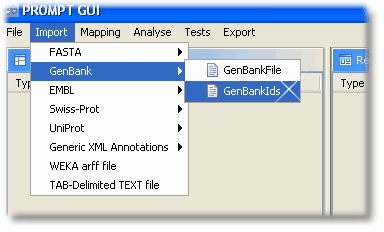
The file with the Swiss-Prot codes can be loaded by choosing "Import -> Swiss-Prot -> SwissProtIds"
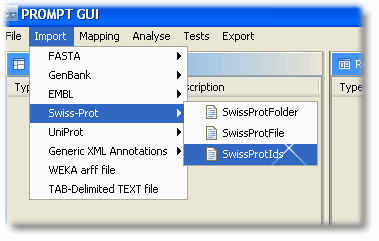
Step 2: Mapping & Results
Select both entries n the input view and choose from the menu:
"Mapping -> BLAST one seq. set against another"
or alternatively you can use the FASTA34 algorithm with the menu command
"Mapping -> FASTA one seq. set against another"
Switch to the message tab to see processing messages. After all sequences have been processed, two new results will show up in the result section.
The result contains the mapped sequences along with some other alignment properties.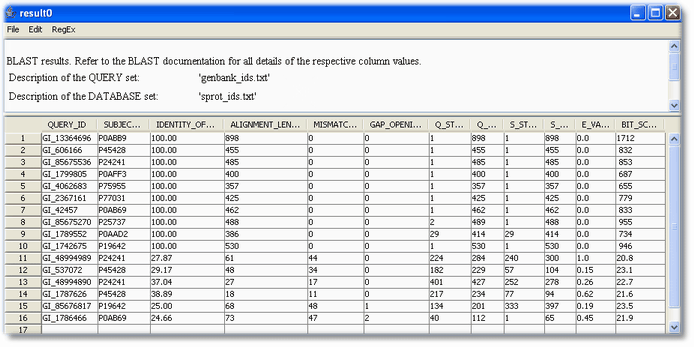
Summary:
- With no effort, you can map identifiers from a database to identifiers from another
- The automatic sequence retrieval in PROMPT makes it even possible to map in cases in which only identifiers and no sequences are available
More:
Start PROMPT, Download PROMPT or sign up to the Community Mailing List
Previous case study: |
Back to the Case studies Overview |
