Case study: Hydrophobicity vs. protein length
View as Movie  BeanShell
script along with data (zipped)
BeanShell
script along with data (zipped) 
Keywords:
computable sequence properties, visualisations (built in and generic), correlation of numerical features
Initial situation:
Assume we want to analyse the membrane proteins of E.coli. Let’s further assume that we have a list of the membrane proteins derived, for example, from using TMHMM (Krogh et al., 2001) (defining all proteins with more than 6 trans-membrane regions as membrane proteins). The other remaining proteins are simply defined as “lysate” proteins.
Questions:
- Is there any relationship between protein length and hydrophobicity i.e. between the GRAVY value (grand average hydrophobicity score) and protein length? Answer this question for the whole genome, membrane and lysate proteins only.
Data:
We have 3 multi FASTA files with amino acid sequences prepared:
| Data file | Content |
| membrane.fasta | contains all membrane proteins of E.coli (all proteins with more than 6 membrane spanning regions predicted by TMHMM 2.0) |
| fullgenome.fasta | all proteins of E.coli |
| lysate.fasta | all proteins but without the membrane proteins as defined above. |
Steps
Step 1: Data import
Simply import all three datasets to PROMPT by using the FASTA import feature. Choose “Import -> FASTA -> File” from PROMPT's menu and choose “protein” as sequence type in the following dialog.
Step 2: Analysis & Results
In this example we analyse whether the hydrophobicity of membrane proteins
(GRAVY) correlates with protein length. First we select the membrane.fasta
object in the Input area. You can select an entry by simply clicking on it.
To select more than one entry or to deselect entries keep the control-key pressed
while clicking at the entries. Now choose from the PROMPT menu
"Analyse -> Computable sequence properties -> Length
vs. Hydrophobicity".
This will show up as a new entry in the result list. This entry contains hydrophobicity
and protein length for each protein. To visualise, just select this result entry
and use the right mouse click to view the context menu. Use the “Visualize”
Menu to run a simple scatter plot (Fig 1A).
It is possible to compare any numeric features with the “Analyse
-> Generic Annotations” option. Select again the membrane.fasta object
in the Input area but choose now the option
“Analyse -> Generic Annotations -> Compare annotations
within 1 set -> Numeric feature correlation”.
In the following dialog choose the two numeric variables that you want to analyze
i.e. here we choose “length” and “HydrophobcityAvg”.
The generic correlation returns two results: CorNumeric1:statvalues and
CorNumeric1:datapairs.
The first result CorNumeric1:statvalues contains the Pearson correlation value, Pearson correlation test p-value and other statistical values. The lower the p-value of the Pearson correlation test, the less likely it is that the observed correlation is by random. To view these values double-click at the CorNumeric1:statvalues entry or use the context popup menu and choose Show data.
Table 1. Results of the correlation test on protein length against hydrophobicity of lysate proteins. The first 2 rows show the Pearson correlation coefficient and the p-value of the correlation test. Additionally the mean, standard deviation and median as well as minimum and maximum of the length and hydrophobicity values are provided.
| FIELD | VALUE |
| Pearson_correlation | cor -0.6911572 |
| Pearson_correlation_test_pvalue | 2.82E-54 |
| setA_Description | "length" |
| setB_Description | "HydrophobicityAvg" |
| setA_mean | 458.8418 |
| setA_std | 148.8976 |
| setA_median | 438 |
| setA_min | 207 |
| setA_max | 1120 |
| setB_mean | 0.763398 |
| setB_std | 0.226708 |
| setB_median | 0.810462 |
| setB_min | -0.03125 |
| setB_max | 1.185057 |
To reproduce the figure below right click on the CorNumeric1:datapairs and choose your desired plotting type.
Tip: Click at the figures below to enlarge them.
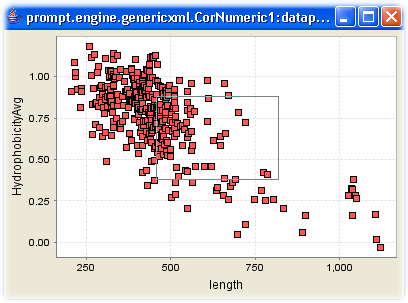
1A. Membrane proteins only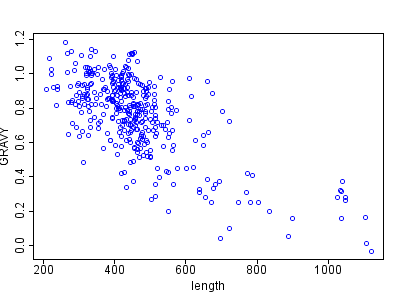 |
1B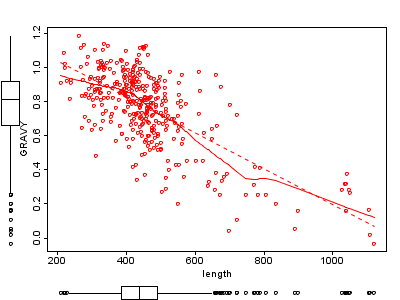 |
Figure 1 A and B: Length of membrane proteins against hydrophobicity (GRAVY). Figure B shows additionally a linear regression line (solid) and a local polynomial loess fitting (dotted line). The generic correlation tests shows a Pearson coefficient of -0.69 with a p-value of 2.8E-54. The blue scatter plot was done with the default R plot method, the red plot uses the car-scatterplot R library.
Additionally PROMPT provides interactive figures that allow one to zoom in and out. Furthermore the actual points can be identified and the accurate figures are shown as tooltips, as seen in Figure 1C.
1C. Interactive scatterplot |
We can repeat this for lysate proteins (Figure 2A) and all proteins (Figure
2B) of E.coli.
2A. Lysate proteins only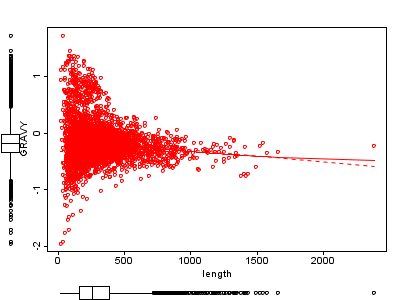 |
2B. Full genome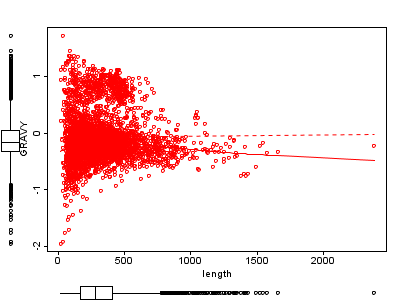 |
Figure 2 Length against hydrophobicity (GRAVY).
A) lysate proteins only, Pearson correlation -0.12 with p-value 1.2E-14
B) all proteins of E.coli, Pearson correlation 0.012 with p-value 0.43
Bottom line of this experiment: Longer membrane proteins tend to be less hydrophobic.
Summary:
- PROMPT can analyse the correlation between any numerical features
- In addition to correlation coefficients like Pearson's correlation, the significance of the observed correlation is computed (here with a Pearson correlation test)
- The graphical plots allow immediate visualisations and further analysis
- As shown in the external data example it is possible to use this type of analysis on any numerical external data.
More:
Start PROMPT, Download PROMPT or sign up to the Community Mailing List
Previous case study: |
Back to the Case studies Overview |
Next case study: Protein abundance analysis in yeast |