Case study: Comparison between thermophilic and mesophilic organisms
View as Movie 
Keywords:
computable sequence properties, visualisations (built in and generic), comparison of numeric distributions
Initial situation:
Complete genomes for mesophilic and thermophilic organism are available and allow one to run large-scale comparisons.
Questions:
- Are proteins of mesophilic and thermophilic proteins longer or shorter or equally long?
- Are thermophilic proteins more hydrophobic than mesophilic proteins?
Data:
We used the proteome sequences of the organisms used by Thompson & Eisenberg (1999) and merged the amino acid sequences to 2 FASTA files.
| File | Content |
| meso_prots.fasta | All proteins of the used mesophilic organisms |
| thermo_prots.fasta | All proteins of the used thermophilic organisms |
Steps
Step 1: Data import
Simply import the 2 protein sets to PROMPT by using the FASTA import feature. Choose “Import -> FASTA -> File” and choose “protein” as sequence type in the following dialog.Step 2: Analysis & Results
Select both input entries and choose from the menu:
"Analyze -> Generic Annotations -> Compare annotations between 2 sets -> Numeric feature comparison"
Then select in both sets the same numerical features that should be compared (here length and HydrophobicityAvg)
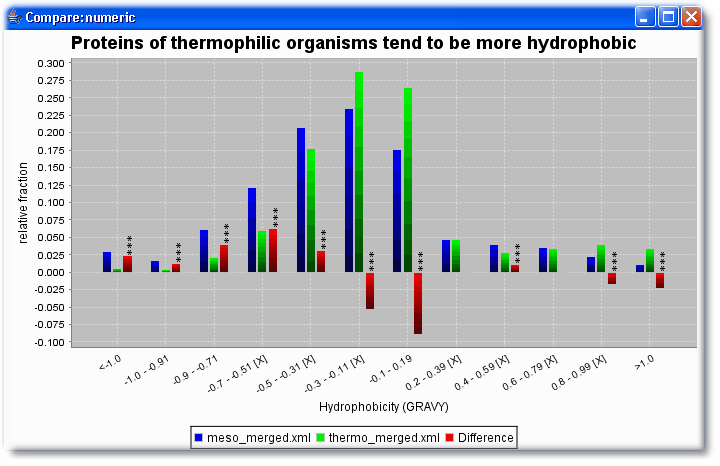
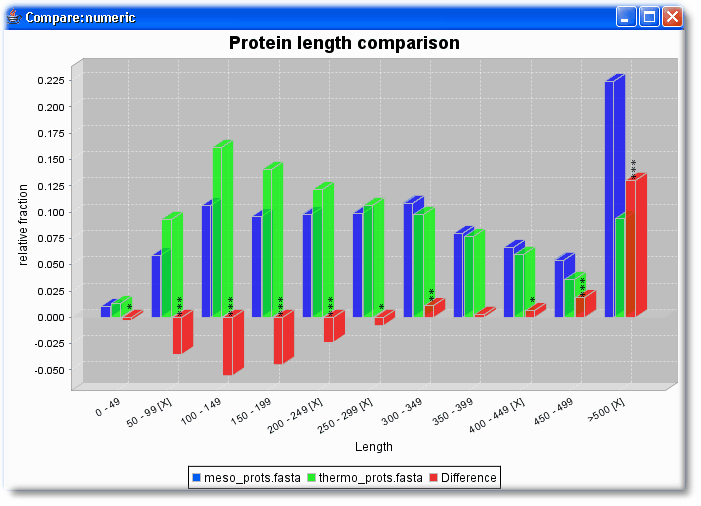
Summary:
- As seen in this example PROMPT can be used to analyse either small sets of proteins or perform large scale analyses corresponding to the 20 organisms in this example.
- All analyses can be run either interactively, by scripts, pipelines or the Java API directly.
More:
Start PROMPT, Download PROMPT or sign up to the Community Mailing List
Previous case study: |
Back to the Case studies Overview |
Next case study: Fast-Mapping against a large remote database like UniProt-Trembl |
